学习决策树
- 概念与计算
- 搭建过程
- 独热码
- 回归树
- 集成学习
- 随机森林
- 提升树
- 与神经网络的比较
- 代码实现
概念
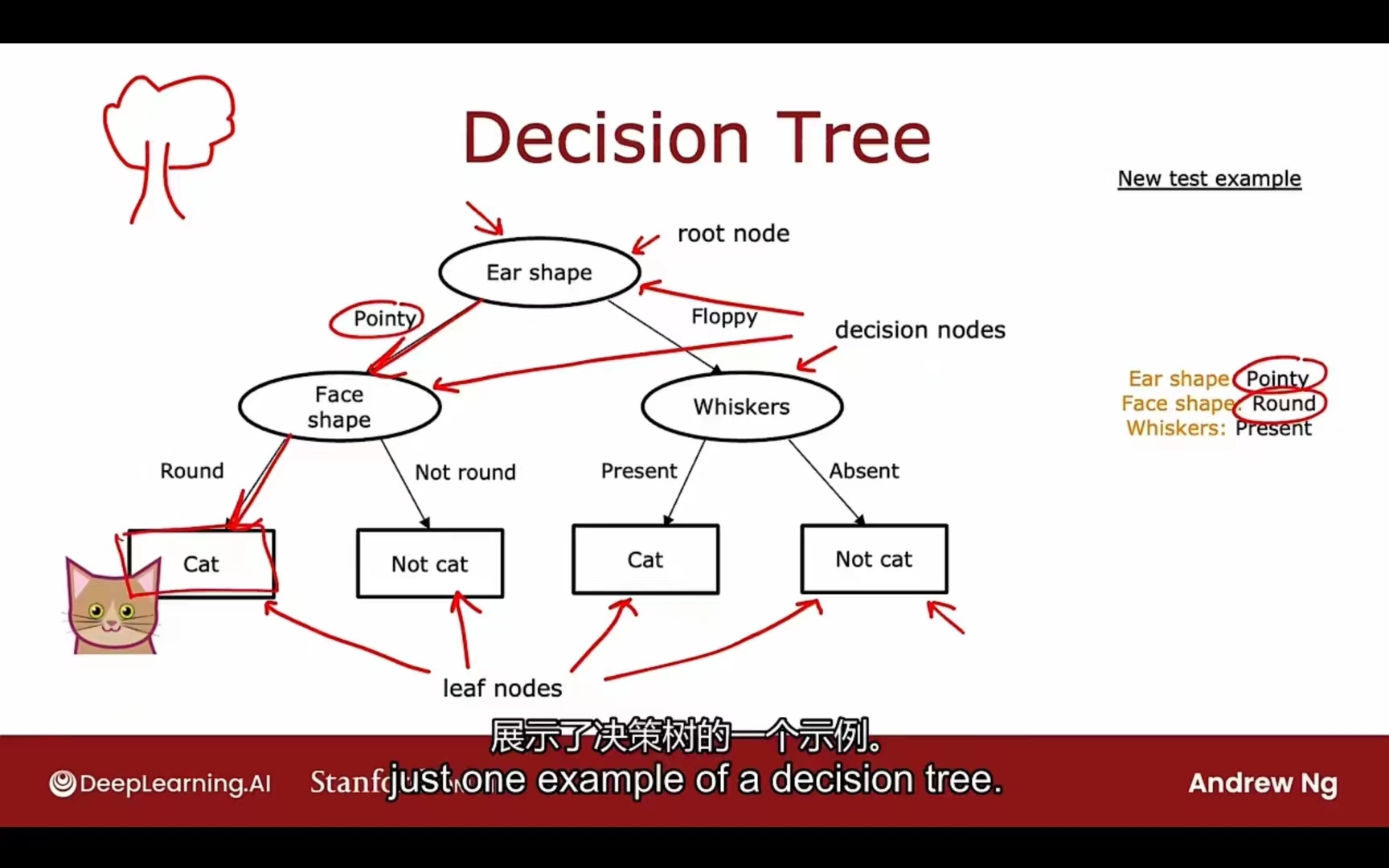
树:
根节点
决策节点
叶节点
决策树学习算法的工作:在所有的可能决策树中尝试选择一个希望在训练集表现良好的情况下,在推广数据集(交叉验证集、测试机)中同样如此的理想决策树模型。
关键点
如何选择哪个特征去区分每个节点
依据最大纯度 -> 熵
什么时候停止划分
- 当一个节点全为1类
- 当划分该节点时会导致树的最大深度增加(防止树变复杂过拟合)
- 纯度分数的提升低于阈值
- 节点中某类的数量低于阈值
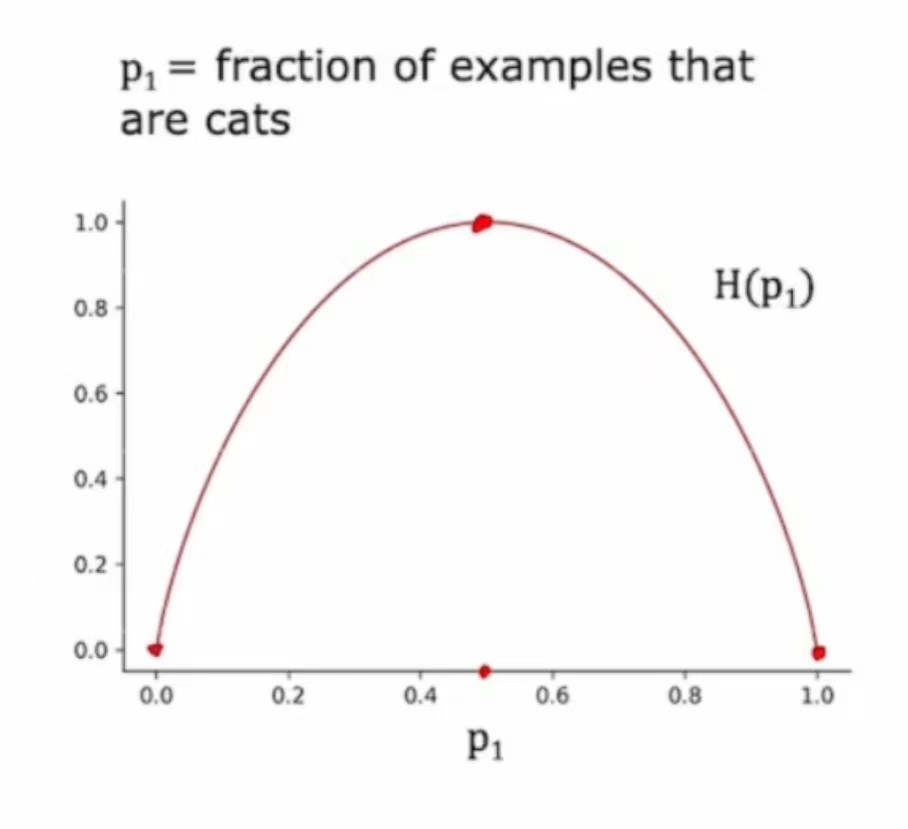
衡量纯度
熵函数:
H(0) = 0
H(1) = 0
H(0.5) = 1
熵的减少被称为信息增益
选择特征的依据:最低加权平均熵
栗子:
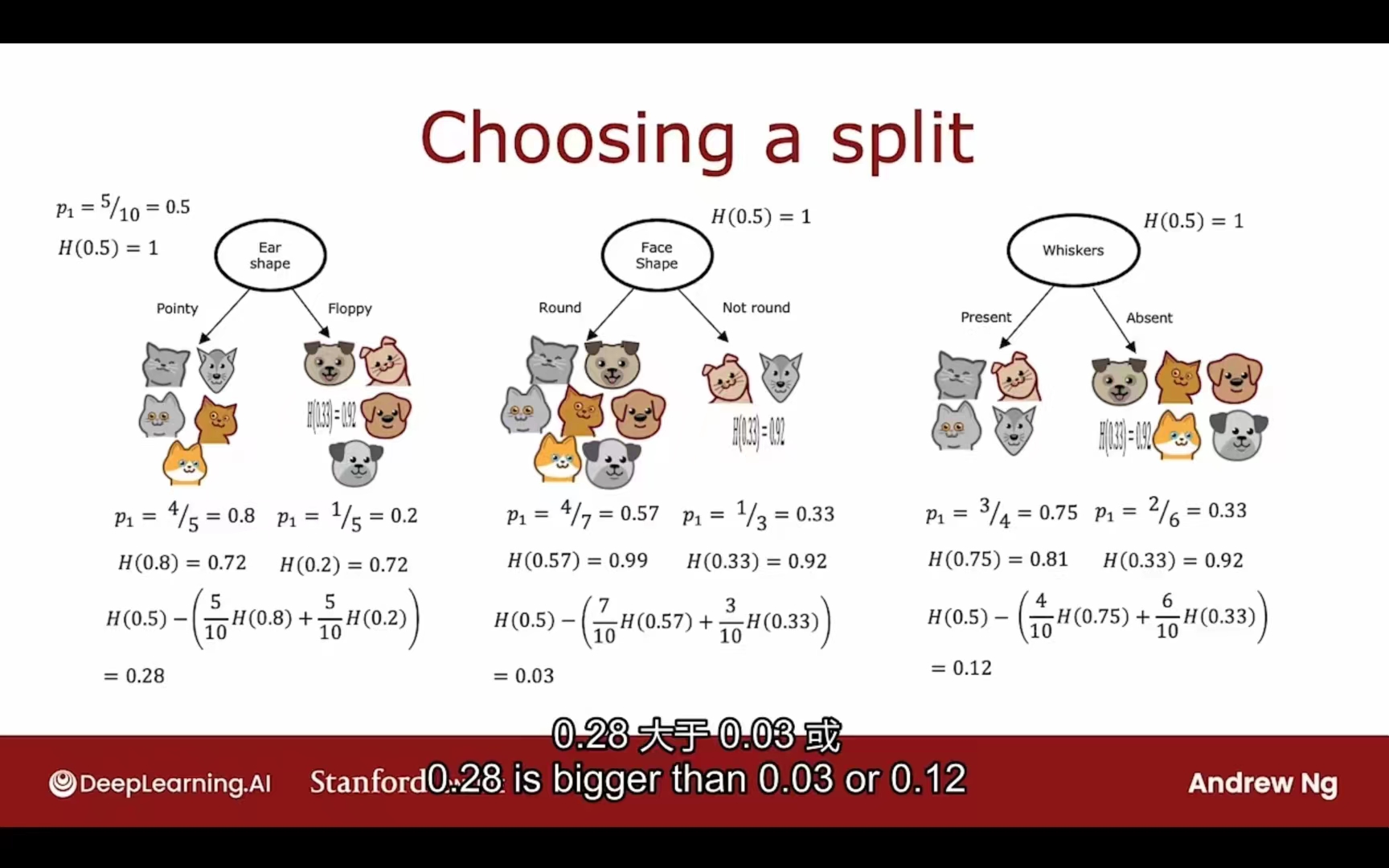
可以给出信息增益的公式:
搭建过程
先序遍历:
- 所有样例从根节点开始
- 分别计算所有潜在特征的信息增益,从中选出最高的
- 根据选中的特征划分数据集,创建树的左右枝干
- 继续划分直到达到停止标准:
- 划分出的数据集纯度100%
- 划分会导致树高超过限制
- 信息增益小于阈值
- 划分子集数量小于阈值
针对连续值特征:
需要考虑每个拆分的标准,根据这个标准计算信息增益,选择信息增益最大的值进行拆分。
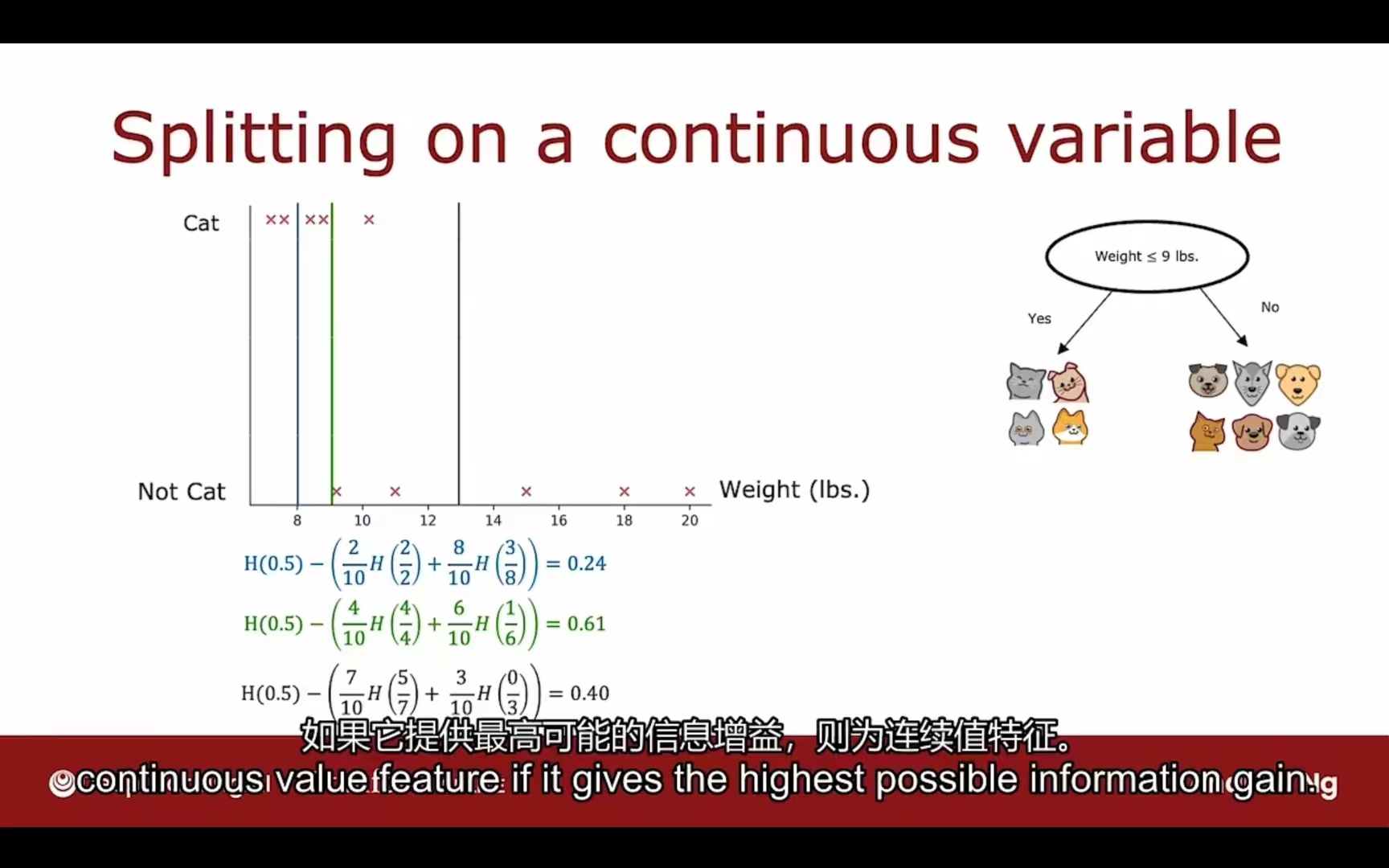
上图选择的具体方法是:在10个数据之间的9个空隙中选择中点,分别计算各自的信息增益,选择最高的那个作为分类依据。
独热码
如果一个分类特征有k种分类方式,那么可以创造k个二进制特征(0 or 1)
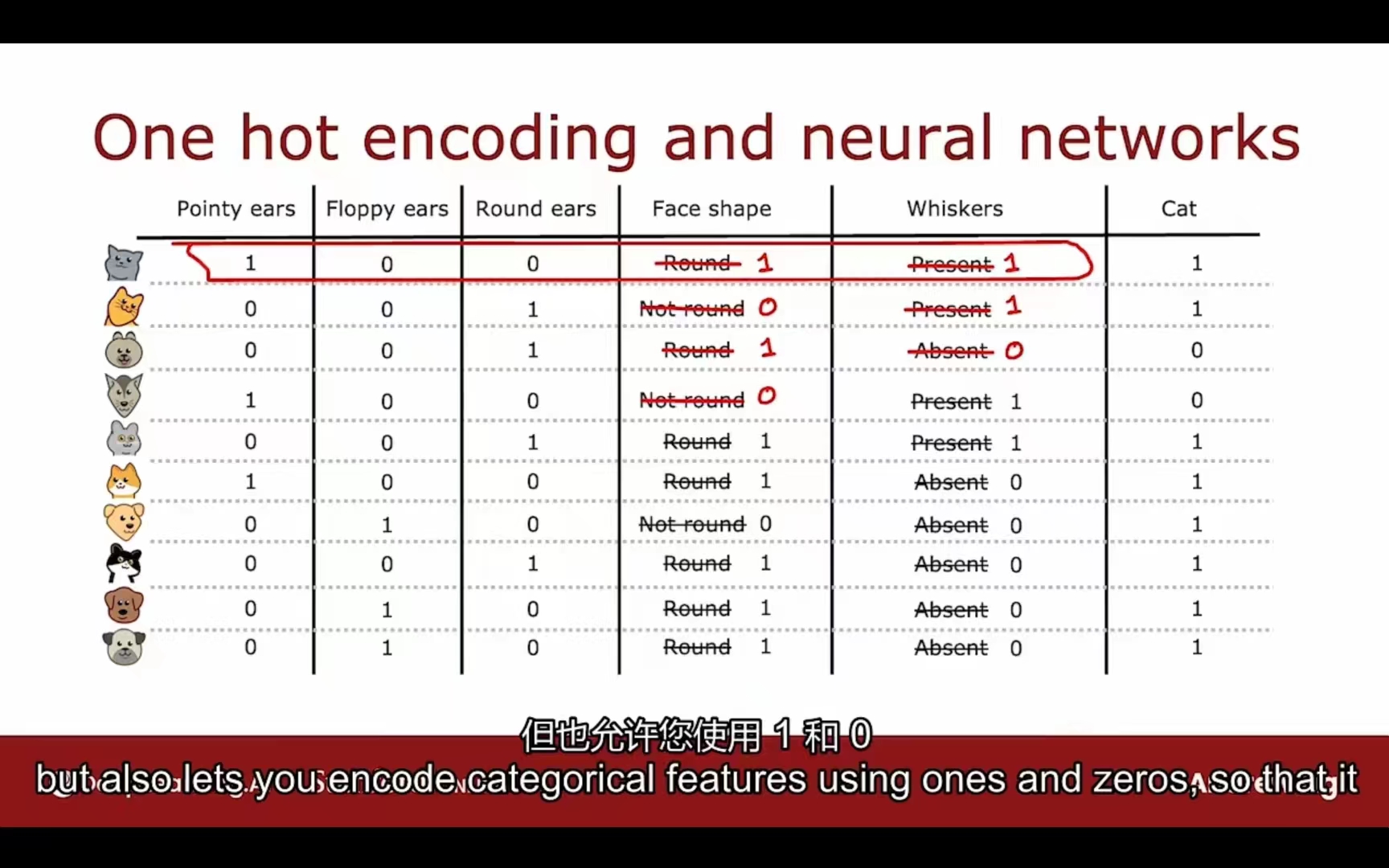
独热码不仅可以用于决策树,也可以用于逻辑回归和神经网络。
注意:独热码只能同时有一个特征为1,其余都为0
回归树
预测数字
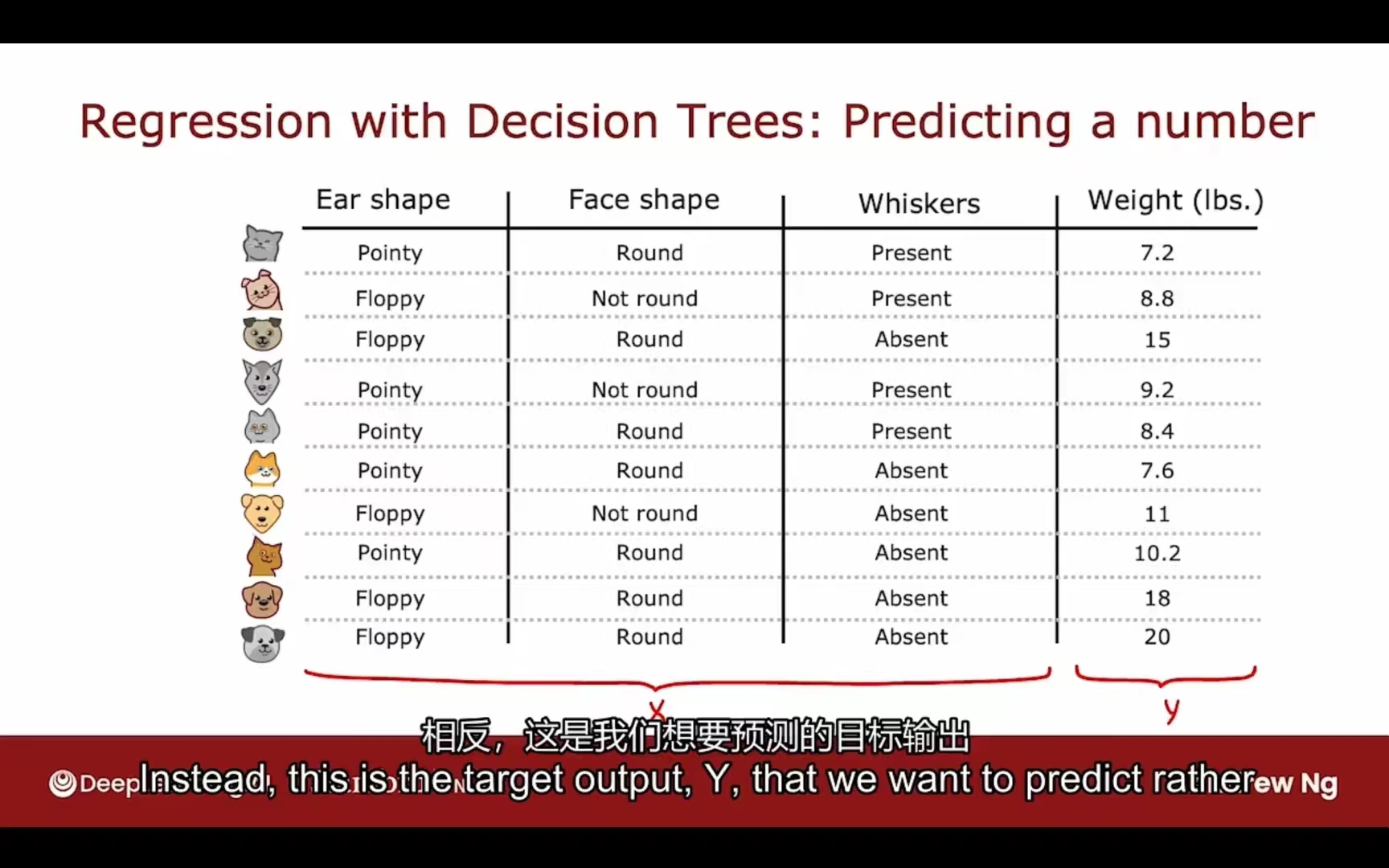
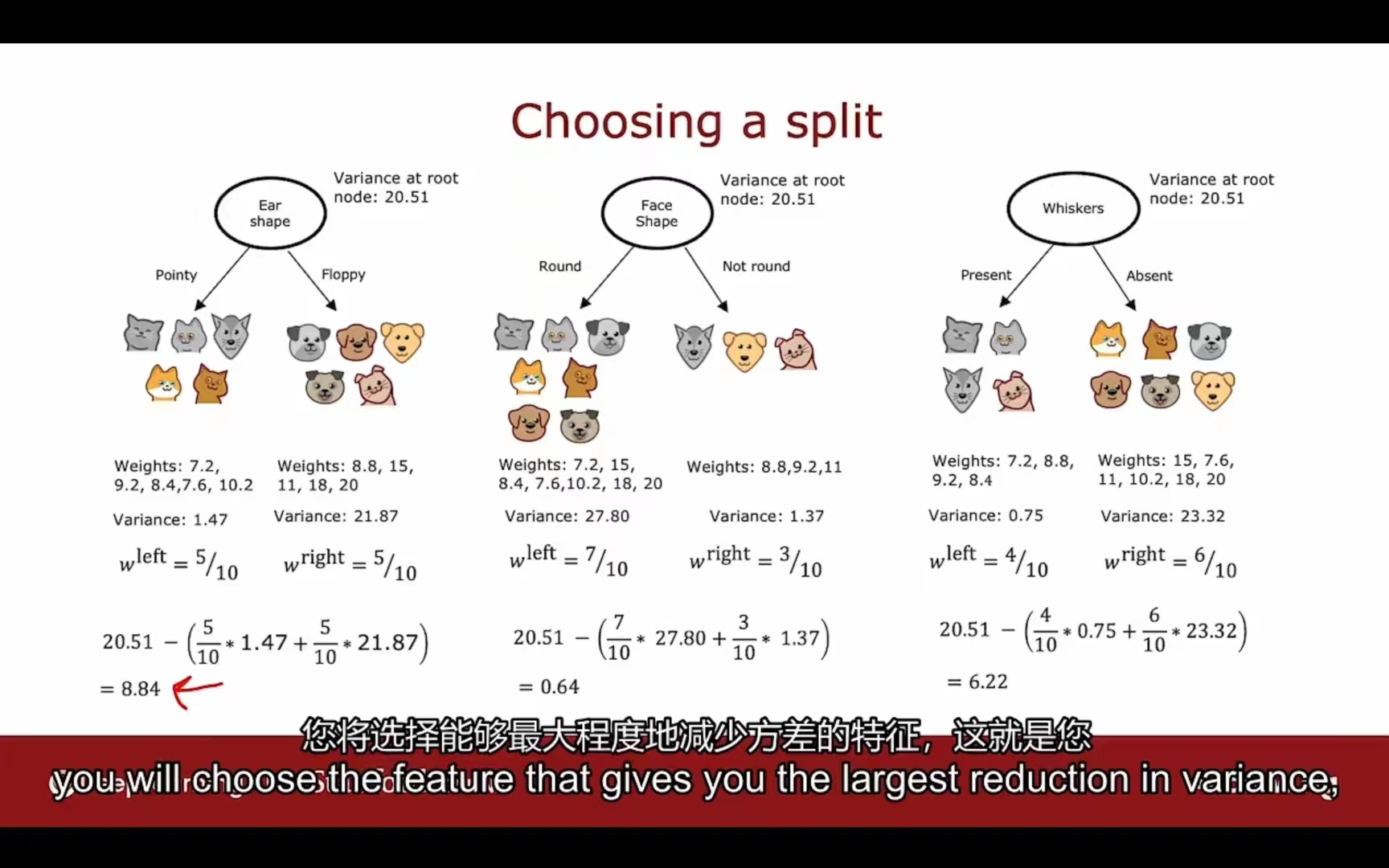
信息增益 -> 测量方差
选择分类特征的过程(类比决策树)
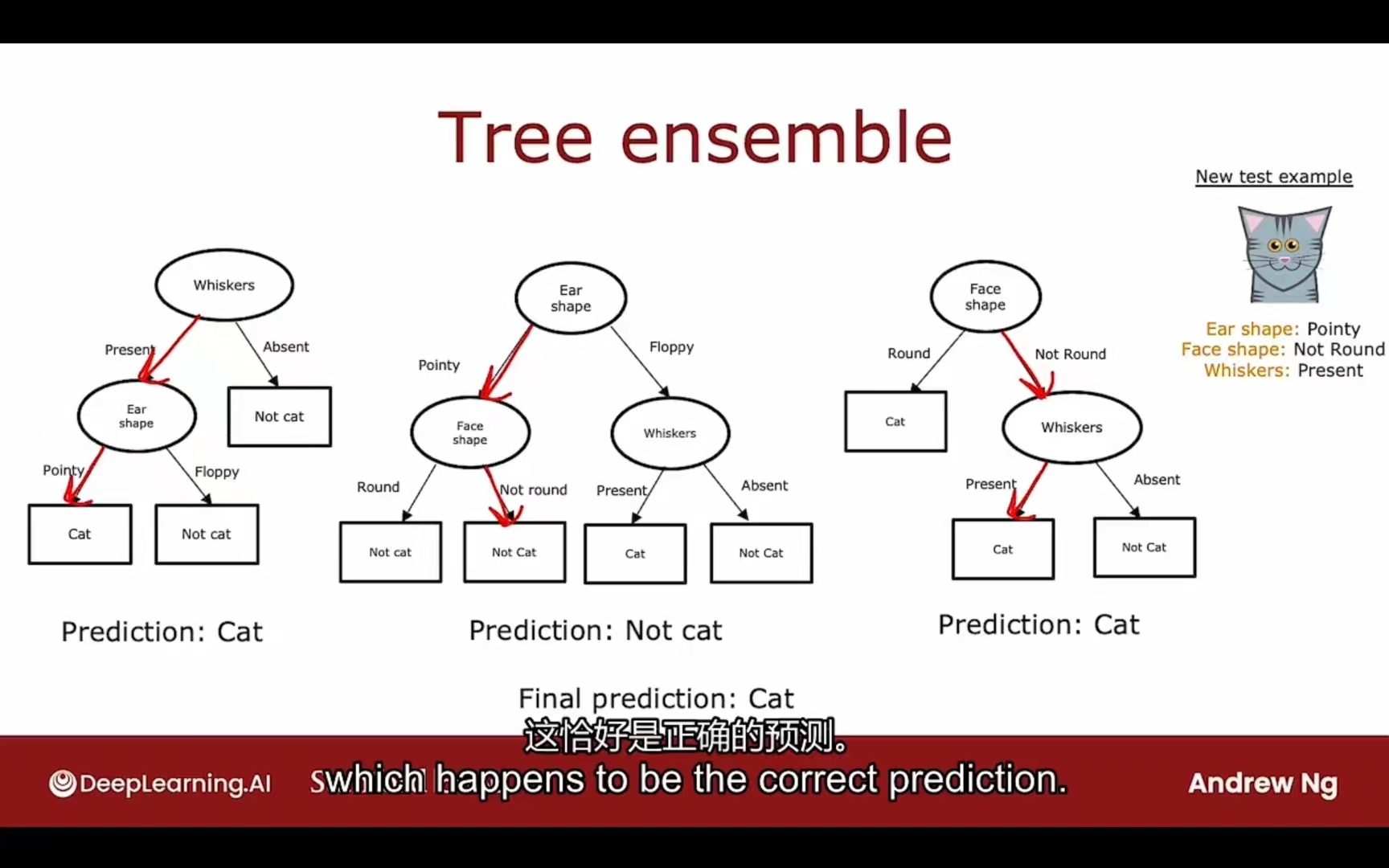
树集合
树对于数据集的改动十分敏感,所以可选择多颗不同类型的决策树加大普适性
随机森林
random forest 集成学习
前提:新的数据集通过有放回抽样进行
随机森林生成过程:
数据集大小:m
1 | for b = 1 to B: # B决定随机森林中子树个数 |
为了使每颗决策子树不同程度高以增加模型普适性,可以采用如下方法:
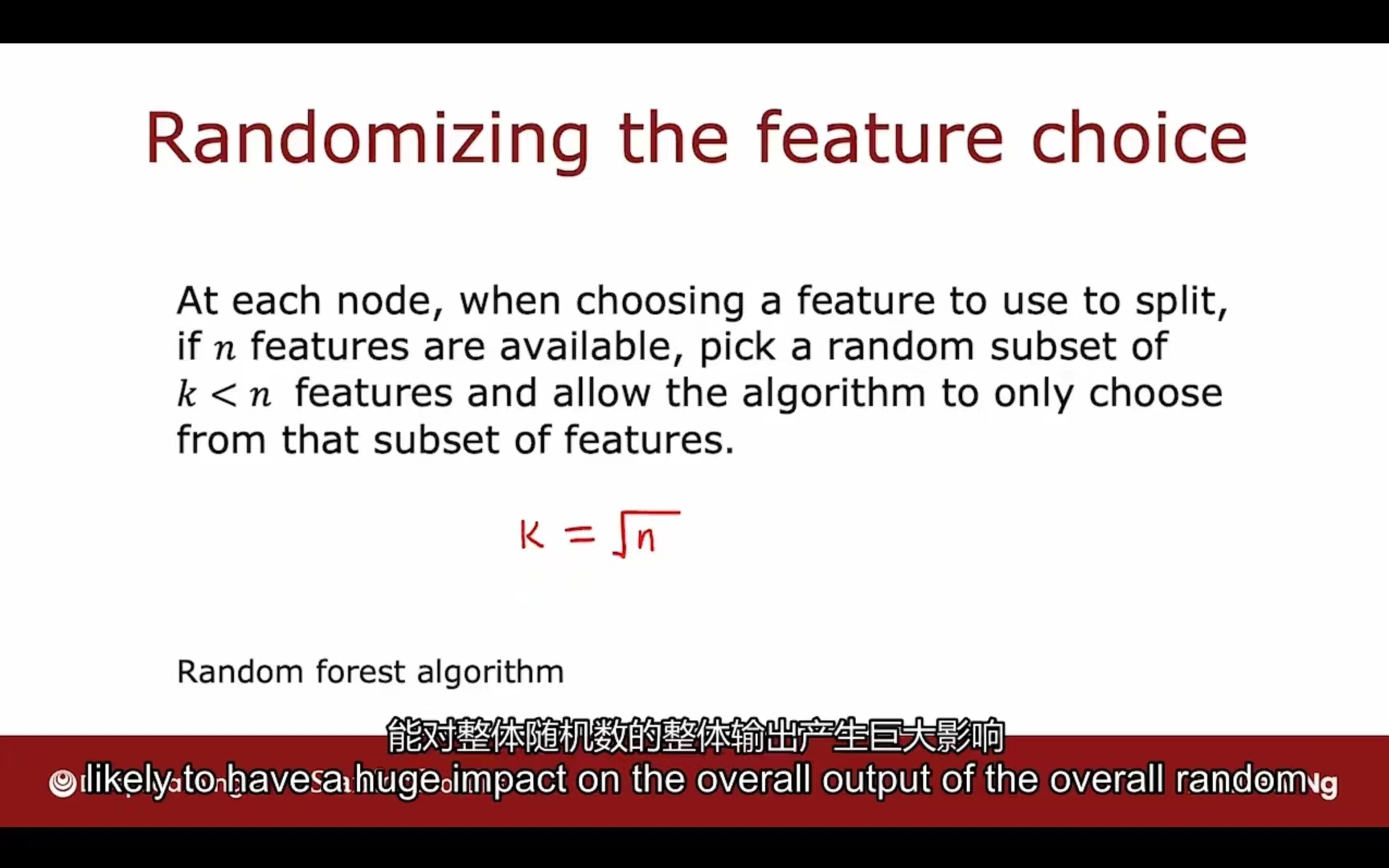
n个特征划分为随机包含其中k个特征的子集,在子集中进行选择
习惯选择$k=\sqrt{n}$
Boosted trees
提升树
1 | for b = 1 to B: # B决定随机森林中子树个数 |
XGBoost
eXtreme Gradient Boosting
- 提升树的开源实现
- 非常快捷和高效
- 有良好的选择默认拆分标准和何时停止拆分的标准
- 内置了正则化以防止过拟合
- 是一种算法竞赛中竞争激烈的算法
分类:
1 | from xgboost import XGBClassifier |
回归:
1 | from xgboost import XGBRegressor |
决策树的使用场景
决策树和树的集成学习:
- 在结构化数据上表现很好(表格等)
- 不推荐在非结构化数据上使用(图像,音频,文字)
- 训练真的很快
- 小型决策树可以人工分析内部
神经网络:
- 在结构化和非结构化数据上都表现很好
- 相对决策树训练会更慢
- 可以和迁移学习一起使用
- 多个神经网络相互独立可以并发训练,而随机森林只能一个接一个训练
代码实现决策树
熵函数:
1 | def compute_entropy(y): |
划分函数:
1 | def split_dataset(X, node_indices, feature): |
信息增益计算函数:
1 | # UNQ_C3 |
找最大信息增益的特征:
1 | def get_best_split(X, y, node_indices): |
核心递归函数建立决策树(先序遍历):
1 | # Not graded |
调用与运行结果展示:
1 | build_tree_recursive(X_train, y_train, root_indices, "Root", max_depth=2, current_depth=0) |